

Welcome to xportr! We have designed xportr
to help get your xpt files ready for transport either to a clinical data
set validator application or to a regulatory agency. This package has
the functionality to associate metadata information to a local R data
frame, perform data set level validation checks and convert into a transport
v5 file(xpt).
As always, we welcome your feedback. If you spot a bug, would like to see a new feature, or if any documentation is unclear - submit an issue on xportr’s GitHub page.
This package is available from CRAN and can be installed by running:
install.packages("xportr")install.packages("xportr", repos = c("https://pharmaverse.r-universe.dev", getOption("repos")))xportr is designed for clinical programmers to create
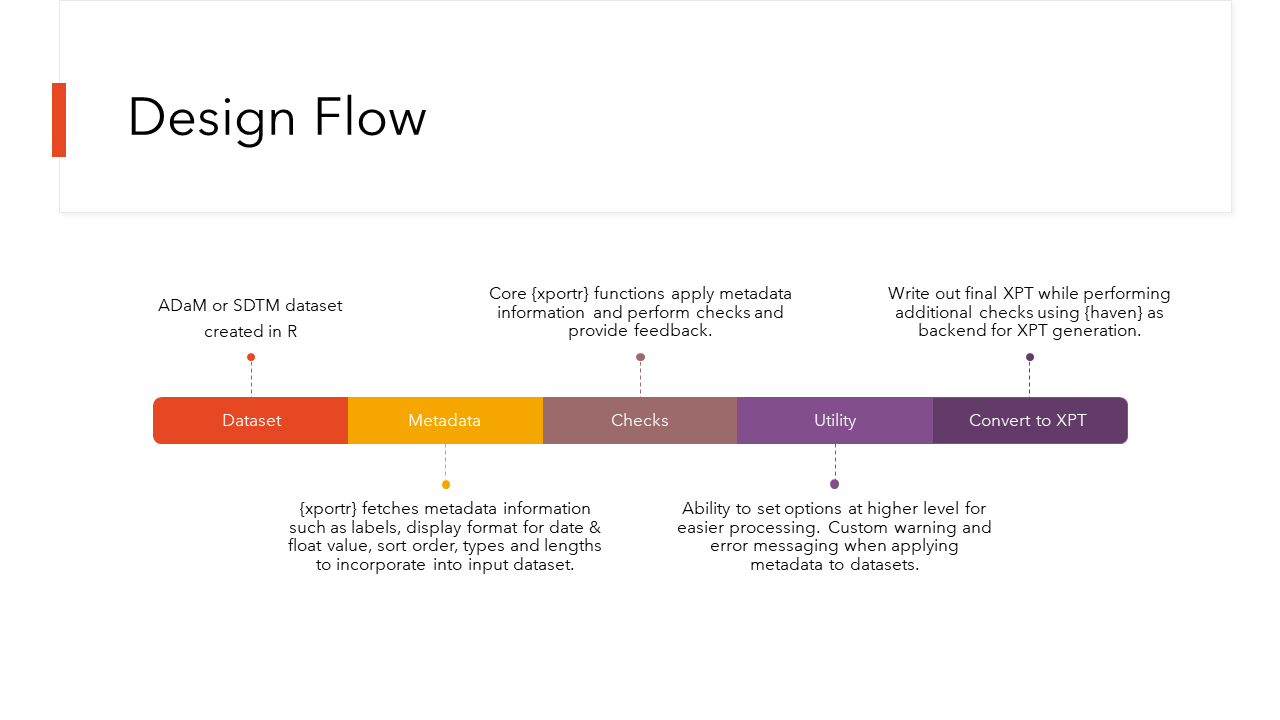
CDISC compliant xpt files- ADaM or
SDTM. Essentially, this package has two big components
to it
The first set of tools are designed to allow a clinical programmer to build a CDISC compliant xpt file directly from R. The second set of tools are to perform checks on your data sets before you send them off to any validators or data reviewers.
NOTE: Each check has associated messages and warning.
Objective: Create a fully compliant v5 xpt
ADSL dataset that was developed using R.
To do this we will need to do the following:
All of which can be done using a well-defined specification file and
the {xportr} package!
First we will start with our ADSL dataset created in R.
This example ADSL dataset contains 306 observations and 51
variables.
library(dplyr)
library(xportr)
data("adsl_xportr")
ADSL <- adsl_xportrWe have created a dummy specification file called
ADaM_spec.xlsx found in the specs folder of
this package. You can use
system.file(file.path("specs/", "ADaM_spec.xlsx"), package = "xportr")
to access this file.
spec_path <- system.file(file.path("specs", "ADaM_spec.xlsx"), package = "xportr")
var_spec <- readxl::read_xlsx(spec_path, sheet = "Variables") %>%
dplyr::rename(type = "Data Type") %>%
dplyr::rename_with(tolower)
dataset_spec <- readxl::read_xlsx(spec_path, sheet = "Datasets") %>%
dplyr::rename(label = "Description") %>%
dplyr::rename_with(tolower)Each xportr_ function has been written in a way to take
in a part of the specification file and apply that piece to the dataset.
Setting verbose = "warn" will send appropriate warning
message to the console. We have suppressed the warning for the sake of
brevity.
ADSL %>%
xportr_metadata(var_spec, "ADSL") %>%
xportr_type(verbose = "warn") %>%
xportr_length(verbose = "warn") %>%
xportr_label(verbose = "warn") %>%
xportr_order(verbose = "warn") %>%
xportr_format() %>%
xportr_df_label(dataset_spec, "ADSL") %>%
xportr_write("adsl.xpt")The xportr_metadata() function can reduce duplication by
setting the variable specification and domain explicitly at the top of a
pipeline. If you would like to use the verbose argument,
you will need to set in each function call.
ADSL %>%
xportr_metadata(var_spec, "ADSL", verbose = "warn") %>%
xportr_type() %>%
xportr_length() %>%
xportr_label() %>%
xportr_order() %>%
xportr_format() %>%
xportr_df_label(dataset_spec) %>%
xportr_write("adsl.xpt")Furthermore, if you’re calling all xportr functions at once with
common metadata and verbosity, you can shorten it by simply using
xportr().
xportr(
.df = ADSL,
var_metadata = var_spec,
df_metadata = dataset_spec,
domain = "ADSL",
verbose = "warn",
"adsl.xpt"
)That’s it! We now have a xpt file created in R with all appropriate
types, lengths, labels, ordering and formats. Please check out the Get
Started for more information and detailed walk through of each
xportr_ function.
We are in talks with other Pharma companies involved with the {pharmaverse} to
enhance this package to play well with other downstream and upstream
packages.