The goal of vmeasur is to quantify the contractile nature of vessels monitored under an operating microscope.
You can install the released version of vmeasur from CRAN with:
install.packages("vmeasur")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("JamesHucklesby/vmeasur")To calibrate an operating microscope, take an image of a gradiated ruler. You can then use this function to calculate the number of pixels per mm.
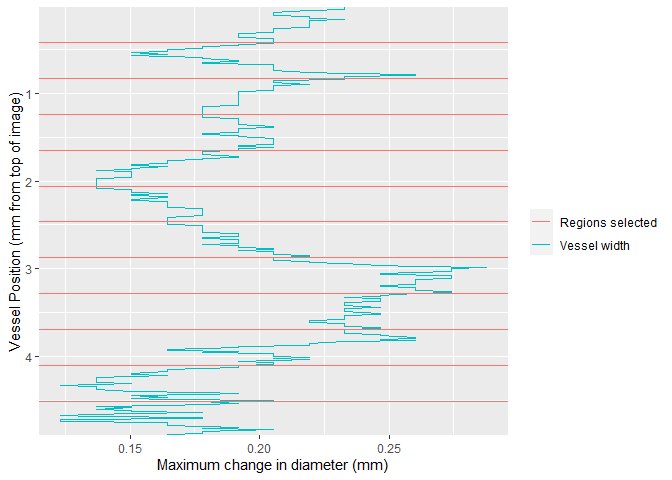
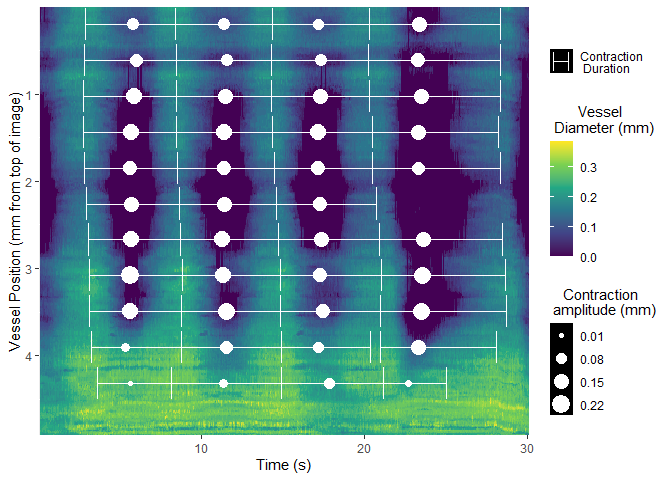
calibrate_pixel_size()Once video data is collected, the region of interest can be selected using select_roi. This provides a wizard that will assist the user through image analysis.
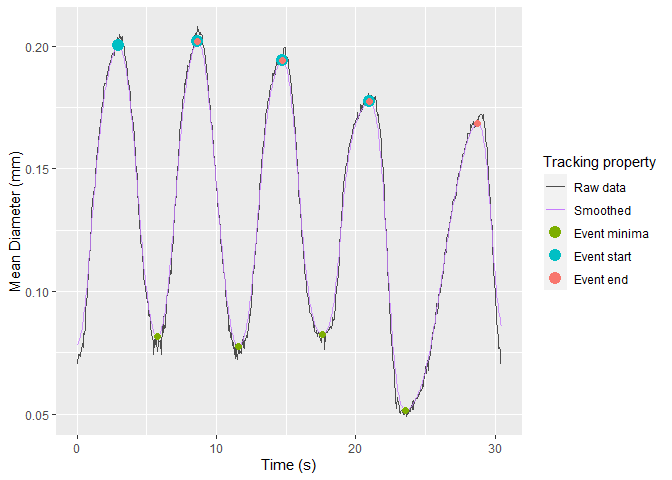
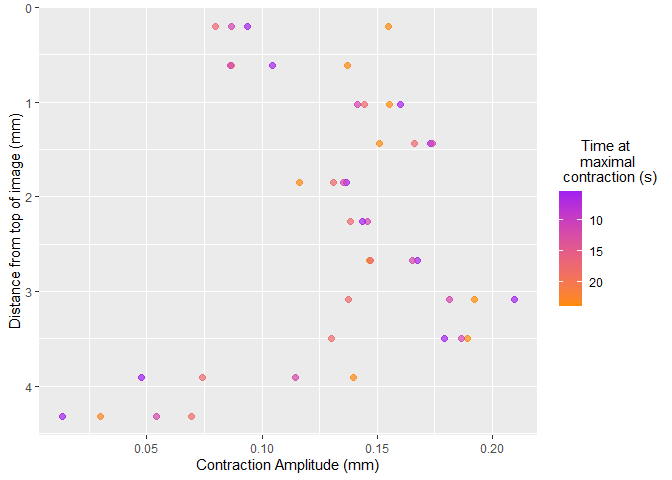
select_roi()Once selected, vmeasur can output a variety of important parameters and graphs

#> # A tibble: 4 x 12
#> # Rowwise:
#> event_maxima event_start event_end type start_value end_value max_value
#> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 531 472 647 contract 12.9 12.3 3.76
#> 2 261 195 331 contract 14.7 14.2 5.67
#> 3 130 67 195 contract 14.6 14.7 5.95
#> 4 396 331 472 contract 14.2 12.9 6.02
#> # ... with 5 more variables: baseline_change <dbl>, event_duration <dbl>,
#> # cont_duration <dbl>, fill_duration <dbl>, event_gradient <dbl>
#> # A tibble: 6 x 4
#> variable mean sd overall
#> <chr> <dbl> <dbl> <chr>
#> 1 CA 0.12 0.00645 0.12 (0.006448)
#> 2 CD 2.77 0.136 2.774 (0.1358)
#> 3 CS 0.0434 0.00390 0.04337 (0.003905)
#> 4 ED 6.36 0.908 6.36 (0.9081)
#> 5 EDD 0.193 0.0112 0.1932 (0.0112)
#> 6 EDD2 0.185 0.0152 0.1854 (0.01517)
#> X.1 y p_width excluded filename
#> 1 1 1 0 FALSE 1
#> 2 2 2 0 FALSE 1
#> 3 3 3 0 FALSE 1
#> 4 4 4 0 FALSE 1
#> 5 5 5 0 FALSE 1
#> 6 6 6 0 FALSE 1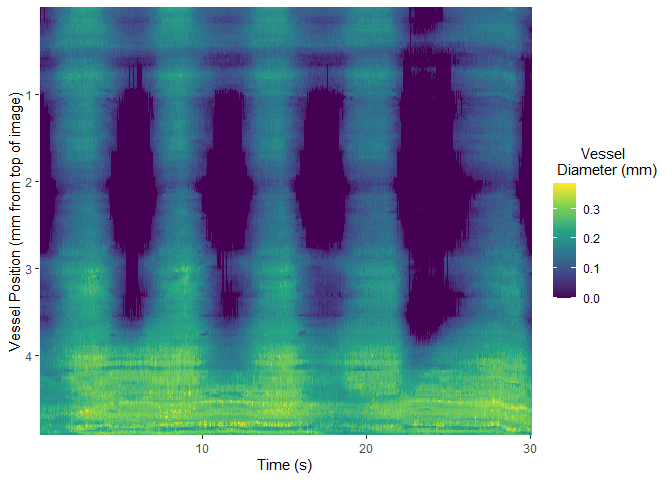
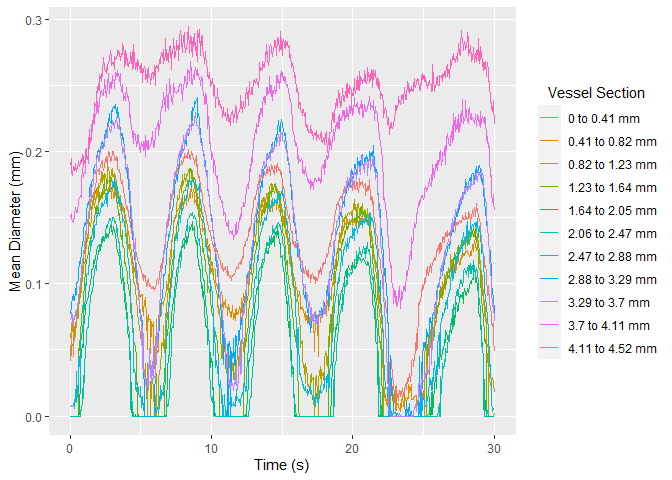
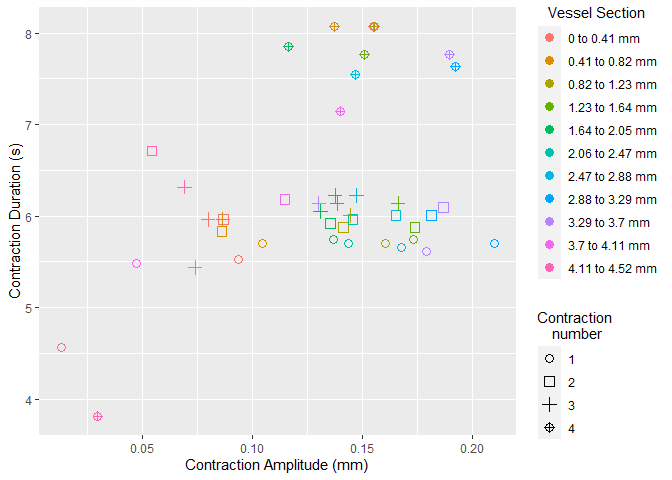
#> # A tibble: 6 x 15
#> # Groups: source_file [6]
#> ygroup event_maxima event_start event_end type start_value end_value
#> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 3 134 62 192 contract 13.2 13.2
#> 2 4 130 62 193 contract 12.6 12.7
#> 3 2 136 63 193 contract 12.3 12.1
#> 4 5 128 63 194 contract 9.97 9.88
#> 5 1 132 65 191 contract 14.2 14.2
#> 6 6 129 66 196 contract 10.5 10.6
#> # ... with 8 more variables: max_value <dbl>, baseline_change <dbl>,
#> # event_duration <dbl>, cont_duration <dbl>, fill_duration <dbl>,
#> # event_gradient <dbl>, source_file <dbl>, cont_id <int>
#> # A tibble: 6 x 5
#> ygroup variable mean sd overall
#> <chr> <chr> <dbl> <dbl> <chr>
#> 1 1 CA 0.104 0.0345 0.1038 (0.03449)
#> 2 1 CD 2.96 0.0913 2.961 (0.0913)
#> 3 1 CS 0.0349 0.0105 0.03486 (0.01051)
#> 4 1 ED 6.38 1.14 6.382 (1.145)
#> 5 1 EDD 0.188 0.00948 0.1875 (0.009478)
#> 6 1 EDD2 0.177 0.0190 0.1768 (0.01904)