The goal of stepcount is to wrap up the https://github.com/OxWearables/stepcount algorithm.
stepcount
Python ModuleSee https://github.com/OxWearables/stepcount?tab=readme-ov-file#install
for how to install the stepcount python module.
In R, you can do this via:
envname = "stepcountblah"
reticulate::conda_create(envname = envname, packages = c("python=3.9", "openjdk", "pip"))
Sys.unsetenv("RETICULATE_PYTHON")
reticulate::use_condaenv(envname)
reticulate::py_install("stepcount", envname = envname, method = "conda", pip = TRUE)Once this is finished, you should be able to check this via:
stepcount::have_stepcount_condaenv()
stepcount::have_stepcount()In some cases, you ay want to set RETICULATE_PYTHON
variable:
stepcount::unset_reticulate_python()
clist = reticulate::conda_list()
Sys.setenv(RETICULATE_PYTHON = clist$python[clist$name == "stepcount"])
stepcount::use_stepcount_condaenv()If you are using the Random Forest model from stepcount,
you may need hmmlearn<0.3.0 due to some issues with its
new implementation of its models as described https://github.com/OxWearables/stepcount/issues/62 (Feb
2024).
stepcount R
PackageYou can install the development version of stepcount
from GitHub with:
# install.packages("devtools")
devtools::install_github("jhuwit/stepcount")stepcount conda environmentIn order to use the stepcount conda environment that
OxWearables recommends, you must run this command before
reticulate is loaded:
stepcount::use_stepcount_condaenv()RETICULATE_PYTHON environment variableIf you have the RETICULATE_PYTHON environment variable
in your .Renviron or your PATH, then
reticulate will still use that version of Python and the
code will likely not work. The unset_reticulate_python()
function will unset that environment variable. So the usage would start
with something like:
stepcount::unset_reticulate_python()
stepcount::use_stepcount_condaenv()and if you need reticulate, you would load it after
stepcount::unset_reticulate_python()
stepcount::use_stepcount_condaenv()
library(reticulate)The stepcount_check function can determine if the
stepcount module can be loaded
stepcount::stepcount_check()
#> [1] TRUEstepcount
(file)The main function is stepcount::stepcount, which takes
can take in a file directly:
library(stepcount)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
library(ggplot2)
library(tidyr)
#> Warning: package 'tidyr' was built under R version 4.3.2
file = system.file("extdata/P30_wrist100.csv.gz", package = "stepcount")
if (stepcount_check()) {
out = stepcount(file = file)
}
#> Loading model...
#> Downloading https://wearables-files.ndph.ox.ac.uk/files/models/stepcount/ssl-20230208.joblib.lzma...
#> Gravity calibration...Gravity calibration... Done! (0.05s)
#> Nonwear detection...Nonwear detection... Done! (0.04s)
#> Resampling...Resampling... Done! (0.05s)
#> Predicting from Model
#> Running step counter...
#> Defining windows...
#> Using local /Users/johnmuschelli/miniconda3/envs/stepcount/lib/python3.9/site-packages/stepcount/torch_hub_cache/OxWearables_ssl-wearables_v1.0.0
#> Classifying windows...
#> Processing ResultLet’s see inside the output, which is a list of values, namely a
data.frame of steps with the time (in 10s
increments) and the number of steps in those 10 seconds, a
data.frame named walking which has indicators
for if there is walking within that 10 second period:
names(out)
#> [1] "steps" "walking" "step_times" "summary"
#> [5] "summary_adjusted" "info"
str(out)
#> List of 6
#> $ steps :'data.frame': 361 obs. of 2 variables:
#> ..$ time : POSIXct[1:361], format: "2019-07-22 14:34:45" "2019-07-22 14:34:55" ...
#> ..$ steps: num [1:361] 0 0 0 0 0 0 0 0 0 0 ...
#> $ walking :'data.frame': 361 obs. of 2 variables:
#> ..$ time : POSIXct[1:361], format: "2019-07-22 14:34:45.88" "2019-07-22 14:34:55.88" ...
#> ..$ walking: num [1:361] 0 0 0 0 0 0 0 0 0 0 ...
#> $ step_times :'data.frame': 5739 obs. of 1 variable:
#> ..$ time: chr [1:5739] "2019-07-22 14:36:26.4899" "2019-07-22 14:36:26.9566" "2019-07-22 14:36:27.4899" "2019-07-22 14:36:27.9566" ...
#> $ summary :List of 16
#> ..$ total : int 5739
#> ..$ hourly : int [1:2(1d)] 2424 3315
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ : chr [1:2] "2019-07-22 14:00:00" "2019-07-22 15:00:00"
#> ..$ daily_stats :'data.frame': 1 obs. of 5 variables:
#> .. ..$ Walk(mins): num 54
#> .. ..$ Steps : num 5739
#> .. ..$ StepsQ1At : chr "14:49:45"
#> .. ..$ StepsQ2At : chr "15:06:45"
#> .. ..$ StepsQ3At : chr "15:20:55"
#> .. ..- attr(*, "pandas.index")=DatetimeIndex(['2019-07-22'], dtype='datetime64[ns]', name='time', freq='D')
#> ..$ daily_avg : int 5739
#> ..$ daily_med : int 5739
#> ..$ daily_min : int 5739
#> ..$ daily_max : int 5739
#> ..$ total_walk : int 54
#> ..$ daily_walk_avg: int 54
#> ..$ daily_walk_med: int 54
#> ..$ daily_walk_min: int 54
#> ..$ daily_walk_max: int 54
#> ..$ cadence_peak1 : int 123
#> ..$ cadence_peak30: int 113
#> ..$ daily_QAt_avg : chr [1:3(1d)] "14:49:45" "15:06:45" "15:20:55"
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ : chr [1:3] "StepsQ1At" "StepsQ2At" "StepsQ3At"
#> ..$ daily_QAt_med : chr [1:3(1d)] "14:49:45" "15:06:45" "15:20:55"
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ : chr [1:3] "StepsQ1At" "StepsQ2At" "StepsQ3At"
#> $ summary_adjusted:List of 16
#> ..$ total : num NaN
#> ..$ hourly : num [1:24(1d)] NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN ...
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ : chr [1:24] "2019-07-22 00:00:00" "2019-07-22 01:00:00" "2019-07-22 02:00:00" "2019-07-22 03:00:00" ...
#> ..$ daily_stats :'data.frame': 1 obs. of 5 variables:
#> .. ..$ Walk(mins): num NaN
#> .. ..$ Steps : num NaN
#> .. ..$ StepsQ1At : num NaN
#> .. ..$ StepsQ2At : num NaN
#> .. ..$ StepsQ3At : num NaN
#> .. ..- attr(*, "pandas.index")=DatetimeIndex(['2019-07-22'], dtype='datetime64[ns]', name='time', freq='D')
#> ..$ daily_avg : num NaN
#> ..$ daily_med : num NaN
#> ..$ daily_min : num NaN
#> ..$ daily_max : num NaN
#> ..$ total_walk : num NaN
#> ..$ daily_walk_avg: num NaN
#> ..$ daily_walk_med: num NaN
#> ..$ daily_walk_min: num NaN
#> ..$ daily_walk_max: num NaN
#> ..$ cadence_peak1 : int 124
#> ..$ cadence_peak30: int 114
#> ..$ daily_QAt_avg : num [1:3(1d)] NaN NaN NaN
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ : chr [1:3] "StepsQ1At" "StepsQ2At" "StepsQ3At"
#> ..$ daily_QAt_med : num [1:3(1d)] NaN NaN NaN
#> .. ..- attr(*, "dimnames")=List of 1
#> .. .. ..$ : chr [1:3] "StepsQ1At" "StepsQ2At" "StepsQ3At"
#> $ info :List of 12
#> ..$ Filename : chr "/Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/library/stepcount/extdata/P30_wrist100.csv.gz"
#> ..$ Device : chr ".csv"
#> ..$ Filesize(MB) : num 4.8
#> ..$ SampleRate : int 100
#> ..$ CalibOK : int 0
#> ..$ CalibErrorBefore(mg) : num NaN
#> ..$ CalibErrorAfter(mg) : num NaN
#> ..$ WearTime(days) : num 0.0417
#> ..$ NonwearTime(days) : num 0
#> ..$ NumNonwearEpisodes : int 0
#> ..$ ResampleRate : int 30
#> ..$ NumTicksAfterResample: int 108001
head(out$steps)
#> time steps
#> 1 2019-07-22 14:34:45 0
#> 2 2019-07-22 14:34:55 0
#> 3 2019-07-22 14:35:05 0
#> 4 2019-07-22 14:35:15 0
#> 5 2019-07-22 14:35:25 0
#> 6 2019-07-22 14:35:35 0
tail(out$steps)
#> time steps
#> 356 2019-07-22 15:33:55 20
#> 357 2019-07-22 15:34:05 20
#> 358 2019-07-22 15:34:15 20
#> 359 2019-07-22 15:34:25 19
#> 360 2019-07-22 15:34:35 17
#> 361 2019-07-22 15:34:45 NaN
tail(out$walking)
#> time walking
#> 356 2019-07-22 15:33:55 1
#> 357 2019-07-22 15:34:05 1
#> 358 2019-07-22 15:34:15 1
#> 359 2019-07-22 15:34:25 1
#> 360 2019-07-22 15:34:35 1
#> 361 2019-07-22 15:34:45 NaNThe step_times data.frame indicates which
times are when steps occurred (at the original sample rate). Make sure
you have the digits.secs option set to see the sub-seconds
for the times (esp for writing out files in
readr::write_csv):
head(out$step_times)
#> time
#> 1 2019-07-22 14:36:26.4899
#> 2 2019-07-22 14:36:26.9566
#> 3 2019-07-22 14:36:27.4899
#> 4 2019-07-22 14:36:27.9566
#> 5 2019-07-22 14:36:30.4566
#> 6 2019-07-22 14:36:30.8233
options(digits.secs = 3)
head(out$step_times)
#> time
#> 1 2019-07-22 14:36:26.4899
#> 2 2019-07-22 14:36:26.9566
#> 3 2019-07-22 14:36:27.4899
#> 4 2019-07-22 14:36:27.9566
#> 5 2019-07-22 14:36:30.4566
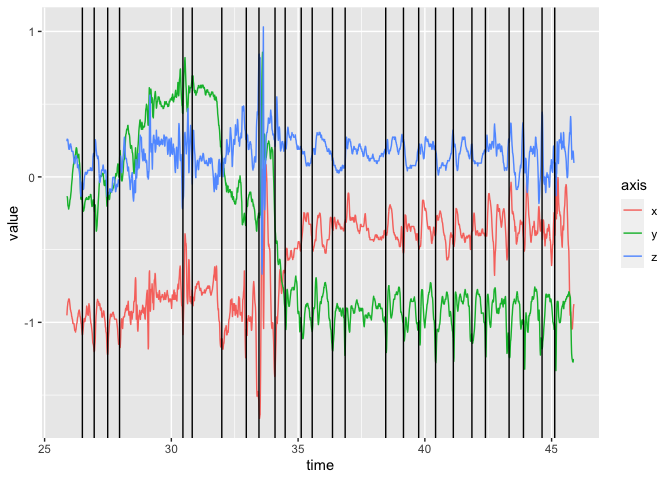
#> 6 2019-07-22 14:36:30.8233We can plot a portion of the tri-axial data and show where the steps were indicated:
df = readr::read_csv(file)
#> Rows: 360001 Columns: 5
#> ── Column specification ────────────────────────────────────────────────────────
#> Delimiter: ","
#> dbl (4): x, y, z, annotation
#> dttm (1): time
#>
#> ℹ Use `spec()` to retrieve the full column specification for this data.
#> ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
if (stepcount_check()) {
st = out$step_times
dat = df[10000:12000,] %>%
dplyr::select(-annotation) %>%
tidyr::gather(axis, value, -time)
st = st %>%
dplyr::mutate(time = lubridate::as_datetime(time)) %>%
dplyr::as_tibble()
st = st %>%
dplyr::filter(time >= min(dat$time) & time <= max(dat$time))
dat %>%
ggplot2::ggplot(ggplot2::aes(x = time, y = value, colour = axis)) +
ggplot2::geom_line() +
ggplot2::geom_vline(data = st, ggplot2::aes(xintercept = time))
}
The main caveat is that stepcount is very precise in the
format of the data, primarily it must have the columns
time, x, y, and z in
the data.
stepcount
(data frame)Alternatively, you can pass out a data.frame, rename the
columns to what you need them to be and then run stepcount
on that:
head(df)
#> # A tibble: 6 × 5
#> time x y z annotation
#> <dttm> <dbl> <dbl> <dbl> <dbl>
#> 1 2019-07-22 14:34:45.890 -0.735 -0.274 -0.481 0
#> 2 2019-07-22 14:34:45.900 -0.591 -0.330 -0.466 0
#> 3 2019-07-22 14:34:45.910 -0.468 -0.496 -0.529 0
#> 4 2019-07-22 14:34:45.920 -0.389 -0.750 -0.670 0
#> 5 2019-07-22 14:34:45.930 -0.369 -1.00 -0.949 0
#> 6 2019-07-22 14:34:45.940 -0.371 -1.17 -1.36 0
out_df = stepcount(file = df)
#> Writing file to CSV...
#> Loading model...
#> Gravity calibration...Gravity calibration... Done! (0.04s)
#> Nonwear detection...Nonwear detection... Done! (0.03s)
#> Resampling...Resampling... Done! (0.06s)
#> Predicting from Model
#> Running step counter...
#> Defining windows...
#> Using local /Users/johnmuschelli/miniconda3/envs/stepcount/lib/python3.9/site-packages/stepcount/torch_hub_cache/OxWearables_ssl-wearables_v1.0.0
#> Classifying windows...
#> Processing ResultWhich gives same output for this data:
all.equal(out[c("steps", "walking", "step_times")],
out_df[c("steps", "walking", "step_times")])
#> [1] TRUEstepcount on multiple filesWhen you pass in multiple files, stepcount will run all
of them, but it will only load the model once, which can have savings,
but the results are still in memory:
if (stepcount_check()) {
out2 = stepcount(file = c(file, file))
length(out2)
names(out2)
# all.equal(out[c("steps", "walking", "step_times")],
# out2[[1]][c("steps", "walking", "step_times")])
}
#> Loading model...
#> Gravity calibration...Gravity calibration... Done! (0.04s)
#> Nonwear detection...Nonwear detection... Done! (0.04s)
#> Resampling...Resampling... Done! (0.06s)
#> Predicting from Model
#> Running step counter...
#> Defining windows...
#> Using local /Users/johnmuschelli/miniconda3/envs/stepcount/lib/python3.9/site-packages/stepcount/torch_hub_cache/OxWearables_ssl-wearables_v1.0.0
#> Classifying windows...
#> Processing Result
#> Gravity calibration...Gravity calibration... Done! (0.04s)
#> Nonwear detection...Nonwear detection... Done! (0.04s)
#> Resampling...Resampling... Done! (0.06s)
#> Predicting from Model
#> Running step counter...
#> Defining windows...
#> Using local /Users/johnmuschelli/miniconda3/envs/stepcount/lib/python3.9/site-packages/stepcount/torch_hub_cache/OxWearables_ssl-wearables_v1.0.0
#> Classifying windows...
#> Processing Result
#> NULLThe model_type parameter indicates the type of model
being run, and the rf will provide the predictions from a
random forest
if (stepcount_check()) {
out_rf = stepcount(file = file, model_type = "rf")
}