

See the pkgdown site at norskregnesentral.github.io/shapr/ for a complete introduction with examples and documentation of the package.
For an overview of the methodology and capabilities of the package
(per shapr v1.0.4), see the software paper Jullum et al.
(2025), available as a preprint here.
With shapr version 1.0.0 (GitHub only, Nov 2024) and
version 1.0.1 (CRAN, Jan 2025), the package underwent a major update,
providing a full restructuring of the code base, and a full suite of new
functionality, including:
explain_forecast() for explaining
forecastsshaprpy making the core functionality of
shapr available in PythonSee the NEWS for a complete list.
shapr version >= 1.0.0 comes with a number of
breaking changes. Most notably, we moved from using two functions
(shapr() and explain()) to one function
(explain()). In addition, custom models are now explained
by passing the prediction function directly to explain().
Several input arguments were renamed, and a few functions for edge cases
were removed to simplify the code base.
Click here to view a version of this README with the old syntax (v0.2.2).
We provide a Python wrapper (shaprpy) which allows
explaining Python models with the methodology implemented in
shapr, directly from Python. The wrapper calls R internally
and therefore requires an installation of R. See here
for installation instructions and examples.
The shapr R package implements an enhanced version of
the Kernel SHAP method for approximating Shapley values, with a strong
focus on conditional Shapley values. The core idea is to remain
completely model-agnostic while offering a variety of methods for
estimating contribution functions, enabling accurate computation of
conditional Shapley values across different feature types, dependencies,
and distributions. The package also includes evaluation metrics to
compare various approaches. With features like parallelized
computations, convergence detection, progress updates, and extensive
plotting options, shapr is a highly efficient and user-friendly tool,
delivering precise estimates of conditional Shapley values, which are
critical for understanding how features truly contribute to
predictions.
A basic example is provided below. Otherwise, we refer to the pkgdown website and the vignettes there for details and further examples.
shapr is available on CRAN and can be
installed in R as:
install.packages("shapr")To install the development version of shapr, available
on GitHub, use
remotes::install_github("NorskRegnesentral/shapr")To also install all dependencies, use
remotes::install_github("NorskRegnesentral/shapr", dependencies = TRUE)shapr supports computation of Shapley values with any
predictive model that takes a set of numeric features and produces a
numeric outcome.
The following example shows how a simple xgboost model
is trained using the airquality dataset, and how
shapr explains the individual predictions.
We first enable parallel computation and progress updates with the following code chunk. These are optional, but recommended for improved performance and user-friendliness, particularly for problems with many features.
# Enable parallel computation
# Requires the future and future_lapply packages
future::plan("multisession", workers = 2) # Increase the number of workers for increased performance with many features
# Enable progress updates of the v(S) computations
# Requires the progressr package
progressr::handlers(global = TRUE)
progressr::handlers("cli") # Using the cli package as backend (recommended for the estimates of the remaining time)Here is the actual example:
library(xgboost)
library(shapr)
data("airquality")
data <- data.table::as.data.table(airquality)
data <- data[complete.cases(data), ]
x_var <- c("Solar.R", "Wind", "Temp", "Month")
y_var <- "Ozone"
ind_x_explain <- 1:6
x_train <- data[-ind_x_explain, ..x_var]
y_train <- data[-ind_x_explain, get(y_var)]
x_explain <- data[ind_x_explain, ..x_var]
# Look at the dependence between the features
cor(x_train)
#> Solar.R Wind Temp Month
#> Solar.R 1.0000000 -0.1243826 0.3333554 -0.0710397
#> Wind -0.1243826 1.0000000 -0.5152133 -0.2013740
#> Temp 0.3333554 -0.5152133 1.0000000 0.3400084
#> Month -0.0710397 -0.2013740 0.3400084 1.0000000
# Fit a basic xgboost model to the training data
model <- xgboost(
data = as.matrix(x_train),
label = y_train,
nround = 20,
verbose = FALSE
)
# Specify phi_0, i.e., the expected prediction without any features
p0 <- mean(y_train)
# Compute Shapley values with Kernel SHAP, accounting for feature dependence using
# the empirical (conditional) distribution approach with bandwidth parameter sigma = 0.1 (default)
explanation <- explain(
model = model,
x_explain = x_explain,
x_train = x_train,
approach = "empirical",
phi0 = p0,
seed = 1
)
#>
#> ── Starting `shapr::explain()` at 2025-08-20 15:08:39 ─────────────────────
#> ℹ Feature classes extracted from the model contains `NA`.
#> Assuming feature classes from the data are correct.
#> ℹ `max_n_coalitions` is `NULL` or larger than or `2^n_features = 16`, and
#> is therefore set to `2^n_features = 16`.
#>
#>
#> ── Explanation overview ──
#>
#>
#>
#> • Model class: <xgb.Booster>
#>
#> • v(S) estimation class: Monte Carlo integration
#>
#> • Approach: empirical
#>
#> • Procedure: Non-iterative
#>
#> • Number of Monte Carlo integration samples: 1000
#>
#> • Number of feature-wise Shapley values: 4
#>
#> • Number of observations to explain: 6
#>
#> • Computations (temporary) saved at:
#> '/tmp/RtmpnBYv2R/shapr_obj_2aa833a1e2267.rds'
#>
#>
#>
#> ── Main computation started ──
#>
#>
#>
#> ℹ Using 16 of 16 coalitions.
# Print the Shapley values for the observations to explain.
print(explanation)
#> explain_id none Solar.R Wind Temp Month
#> <int> <num> <num> <num> <num> <num>
#> 1: 1 43.1 13.212 4.79 -25.6 -5.60
#> 2: 2 43.1 -9.973 5.83 -11.0 -7.83
#> 3: 3 43.1 -2.292 -7.05 -10.2 -4.45
#> 4: 4 43.1 3.325 -3.24 -10.2 -6.66
#> 5: 5 43.1 4.304 -2.63 -14.2 -12.27
#> 6: 6 43.1 0.479 -5.25 -12.6 -6.65
# Provide a formatted summary of the shapr object
summary(explanation)
#>
#> ── Summary of Shapley value explanation ───────────────────────────────────
#> • Computed with`shapr::explain()` in 2.2 seconds, started 2025-08-20
#> 15:08:39
#> • Model class: <xgb.Booster>
#> • v(S) estimation class: Monte Carlo integration
#> • Approach: empirical
#> • Procedure: Non-iterative
#> • Number of Monte Carlo integration samples: 1000
#> • Number of feature-wise Shapley values: 4
#> • Number of observations to explain: 6
#> • Number of coalitions used: 16 (of total 16)
#> • Computations (temporary) saved at:
#> '/tmp/RtmpnBYv2R/shapr_obj_2aa833a1e2267.rds'
#>
#> ── Estimated Shapley values
#> explain_id none Solar.R Wind Temp Month
#> <int> <char> <char> <char> <char> <char>
#> 1: 1 43.09 13.21 4.79 -25.57 -5.60
#> 2: 2 43.09 -9.97 5.83 -11.04 -7.83
#> 3: 3 43.09 -2.29 -7.05 -10.15 -4.45
#> 4: 4 43.09 3.33 -3.24 -10.22 -6.66
#> 5: 5 43.09 4.30 -2.63 -14.15 -12.27
#> 6: 6 43.09 0.48 -5.25 -12.55 -6.65
#> ── Estimated MSEv
#> Estimated MSE of v(S) = 144 (with sd = 64)
# Finally, we plot the resulting explanations
plot(explanation)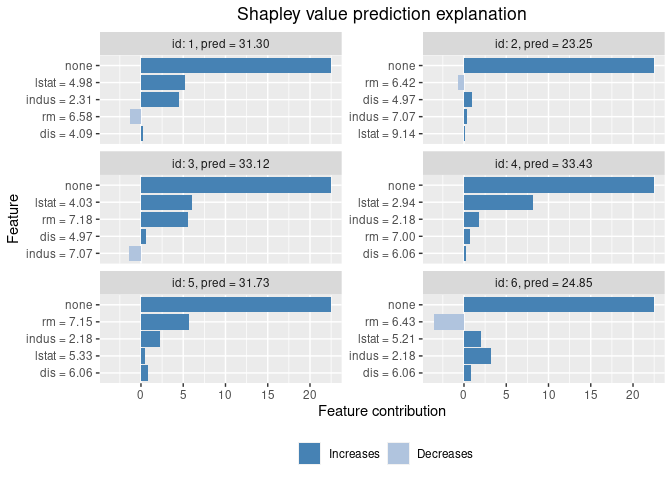
See Jullum et al. (2025) (preprint available here) for a software paper with an overview of the methodology and capabilities of the package (as of v1.0.4). See the general usage vignette for further basic usage examples and brief introductions to the methodology. For more thorough information about the underlying methodology, see methodological papers Aas, Jullum, and Løland (2021), Redelmeier, Jullum, and Aas (2020), Jullum, Redelmeier, and Aas (2021), Olsen et al. (2022), Olsen et al. (2024). See also Sellereite and Jullum (2019) for a very brief paper about a previous version (v0.1.1) of the package (with a different structure, syntax, and significantly less functionality).
All feedback and suggestions are very welcome. Details on how to contribute can be found here. If you have any questions or comments, feel free to open an issue here.
Please note that the shapr project is released with a Contributor
Code of Conduct. By contributing to this project, you agree to abide
by its terms.