The goal of gregRy is to make the GREGORY estimator
easily available to use.
The development version of gregRy is available from GitHub with:
# install.packages("devtools")
devtools::install_github("olekwojcik/gregRy")The package gregRy does not contain a dataset, which is
why our example utilizes the package pdxTrees
library(gregRy)
set.seed(13)
# load and wrangle data
# Overall dataset to create estimates with
# Includes response variable and predictors
family_filter_data <- get_pdxTrees_parks() %>%
as.data.frame() %>%
drop_na(DBH, Crown_Width_NS, Tree_Height) %>%
filter(Condition != "Dead") %>%
select(UserID, Tree_Height, Crown_Width_NS, DBH, Condition, Family) %>%
group_by(Family) %>%
summarize(count = n())
dat <- get_pdxTrees_parks() %>%
as.data.frame() %>%
drop_na(DBH, Crown_Width_NS, Tree_Height) %>%
filter(Condition != "Dead") %>%
select(UserID, Tree_Height, Crown_Width_NS, DBH, Condition, Family) %>%
left_join(family_filter_data, by = "Family") %>%
filter(count > 4) %>%
select(UserID, Tree_Height, Crown_Width_NS, DBH, Condition, Family)
dat_s <- get_pdxTrees_parks() %>%
as.data.frame() %>%
drop_na(DBH, Crown_Width_NS, Tree_Height) %>%
filter(Condition != "Dead") %>%
select(UserID, Tree_Height, Crown_Width_NS, DBH, Condition, Family) %>%
left_join(family_filter_data, by = "Family") %>%
filter(count > 4) %>%
select(UserID, Tree_Height, Crown_Width_NS, DBH, Condition, Family) %>%
group_by(Family) %>%
slice_sample(prop = 0.25) %>%
ungroup()
dat_est <- dat %>%
filter(Family == "Pinaceae")
predictors <- c("Crown_Width_NS", "DBH")
dat_x_bar <- dat %>%
group_by(Family) %>%
summarize(across(predictors,
mean)) %>%
pivot_longer(!Family,
names_to = "variable",
values_to = "mean")
dat_count_est <- dat %>%
group_by(Family) %>%
summarize(count = n())
# Create dataset of proportions using estimation and resolution
dat_prop <- left_join(dat, dat_count_est, by = "Family") %>%
group_by(Condition, Family) %>%
summarize(prop = n()/count) %>%
distinct() %>%
ungroup()
# Create dataset of means of 'pixel' data
dat_x_means <- get_pdxTrees_parks() %>%
as.data.frame() %>%
drop_na(DBH, Crown_Width_NS, Tree_Height) %>%
dplyr::summarize(DBH = mean(DBH), Crown_Width_NS = mean(Crown_Width_NS),
Tree_Height = mean(Tree_Height))
dat_x_bar_new <- dat_x_bar %>%
filter(variable == "Crown_Width_NS") %>%
mutate(Crown_Width_NS = mean) %>%
select(Family, Crown_Width_NS)To use GREGORY, we need 3 different datasets.
The first dataset is the overall data:
#> UserID Tree_Height Crown_Width_NS DBH Condition Family
#> 1 1 105 44 37.4 Fair Pinaceae
#> 2 2 94 49 32.5 Fair Pinaceae
#> 3 3 23 28 9.7 Fair Rosaceae
#> 4 4 28 38 10.3 Poor Fagaceae
#> 5 5 102 43 33.2 Fair Pinaceae
#> 6 6 95 35 32.1 Fair PinaceaeThe second dataset is the means of the predictors at the estimation level (Family estimates):
#> Family Crown_Width_NS
#> 1 Altingiaceae 40.60444
#> 2 Anacardiaceae 19.25000
#> 3 Aquifoliaceae 14.85417
#> 4 Betulaceae 31.08437
#> 5 Bignoniaceae 31.54167
#> 6 Cannabaceae 35.12500The third dataset is contains both the resolution and estimation, with the proportion of resolution in the given estimation unit:
#> Condition Family prop
#> 1 Fair Altingiaceae 0.9066667
#> 2 Fair Anacardiaceae 0.7500000
#> 3 Fair Aquifoliaceae 0.8750000
#> 4 Fair Betulaceae 0.8298755
#> 5 Fair Bignoniaceae 0.7916667
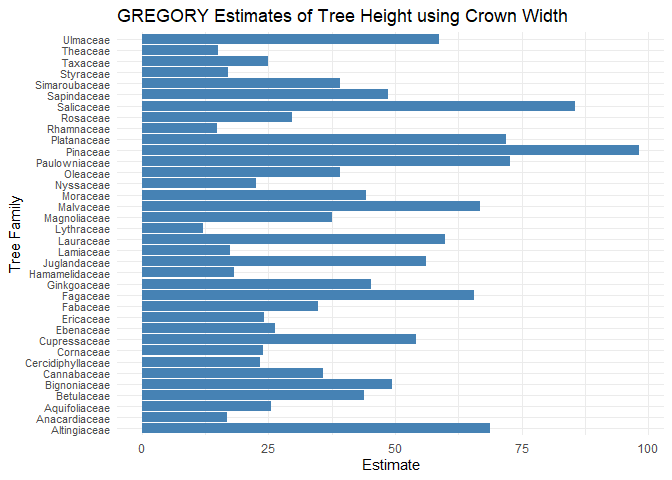
#> 6 Fair Cannabaceae 0.8125000# Create GREGORY estimates
x1 <- gregory_all(plot_df = dat_s %>% drop_na(),
resolution = "Condition",
estimation = "Family",
pixel_estimation_means = dat_x_bar_new,
proportions = dat_prop,
formula = Tree_Height ~ Crown_Width_NS,
prop = "prop")
print(x1)
#> # A tibble: 36 x 3
#> Family estimate variance
#> <chr> <dbl> <dbl>
#> 1 Altingiaceae 68.8 604.
#> 2 Anacardiaceae 16.8 41.3
#> 3 Aquifoliaceae 25.6 73.1
#> 4 Betulaceae 43.9 393.
#> 5 Bignoniaceae 49.4 1277.
#> 6 Cannabaceae 35.9 193.
#> 7 Cercidiphyllaceae 23.3 48.6
#> 8 Cornaceae 23.9 207.
#> 9 Cupressaceae 54.1 1118.
#> 10 Ebenaceae 26.3 NA
#> # ... with 26 more rows
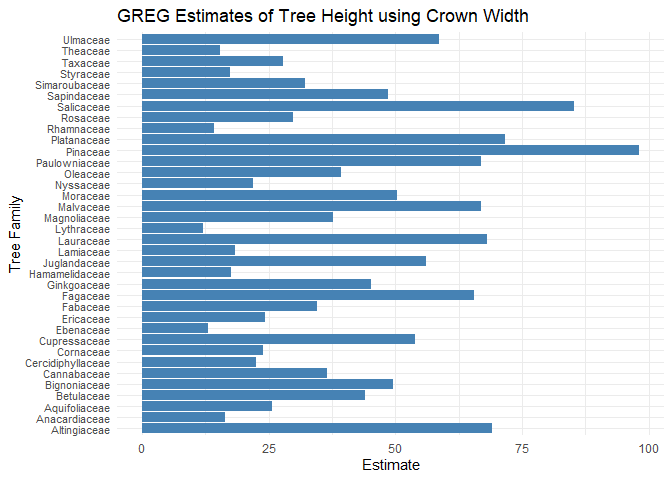
# Create GREG estimates
x2 <- greg_all(plot_df = dat_s %>% drop_na(),
estimation = "Family",
pixel_estimation_means = dat_x_bar_new,
formula = Tree_Height ~ Crown_Width_NS)
print(x2)
#> # A tibble: 36 x 3
#> Family estimate variance
#> <chr> <dbl> <dbl>
#> 1 Altingiaceae 69.0 604.
#> 2 Anacardiaceae 16.5 41.3
#> 3 Aquifoliaceae 25.6 73.1
#> 4 Betulaceae 44.0 393.
#> 5 Bignoniaceae 49.6 1277.
#> 6 Cannabaceae 36.6 193.
#> 7 Cercidiphyllaceae 22.6 48.6
#> 8 Cornaceae 23.9 207.
#> 9 Cupressaceae 53.9 1118.
#> 10 Ebenaceae 13 NA
#> # ... with 26 more rows