
Faster raster processing in R using
GRASS
fasterRaster is an R package designed
specifically to handle large-in-memory/large-on-disk spatial rasters and
vectors. fasterRaster does this using Open Source
Geospatial’s
GRASS
fasterRaster was created with five design
principles:
fasterRaster complements
terra and sf, and is highly dependent on them!
It is useful for analyzing large-in-memory/large-on-disk rasters and
vectors that those packages struggle to handle. For medium- and
small-size objects, terra and sf will almost
always be faster.terra, you basically know how to use
fasterRaster! That’s because most of the functions have the
same name and almost the same arguments as terra
functions.fasterRaster are the same as those from functions in
terra with the same name.GRASS requires users to
track things like “locations” or “projects”, “mapsets”, and “regions”
for which there is no comparable analog in the terra or
sf packages. fasterRaster handles these behind
the scenes so you don’t need to.rgrass package provides a
powerful conduit through which you can run GRASS tools from
R. As such, it provides much more flexibility than
fasterRaster. However, to use rgrass, you need
to know what GRASS tools you want to use and be familiar
with GRASS syntax. fasterRaster obviates this
step but uses rgrass as a backend, allowing you to focus on
R syntax and look up help for functions the normal way you
do in R. You don’t need to know GRASS!fasterRaster makes heavy use of the
rgrass
package by Roger Bivand and others, the
terra
package by Robert Hijmans, the
sf
package by Edzer Pebesma, Roger Bivand, and others, and of course
GRASS, so is greatly
indebted to all of these creators!
fasterRaster comes with four user-oriented
vignettes, plus a pkgdown
site
with full documentation:
o
Getting
started (also reproduced below)
o
Types
of GRasters
o
Making
fasterRaster faster
o
Addons
o
Documentation
To install fasterRaster, please use:
install_packages('fasterRaster', dependencies = TRUE)
You can get the latest stable release using:
remotes::install_github('adamlilith/fasterRaster', dependencies = TRUE)
…and the development version from:
remotes::install_github('adamlilith/fasterRaster@intuitive_fasterRaster', dependencies = TRUE)
To use fasterRaster you must install GRASS version 8.3+ on your operating
system. You will need to use the stand-alone installer, not the
Open Source Geospatial (OS Geo) installer.
Optional: A few functions in
fasterRaster require GRASS “addon”
tools, which do not come bundled with GRASS. You do not
need to install these addons if you do not use functions that call them.
A list of functions that require addons can be seen in the “addons”
vignette (in R, use
vignette("addons", package = "fasterRaster")). This
vignette also explains how to install addons.
The example presented here is the same as that presented in the the “getting started” vignette.
We’ll do a simple operation in which we:
Add a buffer to lines representing rivers, then
Calculate the distance to from each cell to the closest buffer and burn the distance values into a raster.
To do this, we’ll be using maps representing the middle of the
eastern coast of Madagascar. We will also use the terra and
sf packages.
library(terra) # GIS for rasters and vectors
library(sf) # GIS for vectors
library(fasterRaster)
# Get example elevation raster and rivers vector:
madElev <- fastData('madElev') # SpatRaster with elevation
madRivers <- fastData('madRivers') # sp vector with rivers
# Plot inputs:
plot(madElev)
plot(st_geometry(madRivers), col = "lightblue", add = TRUE)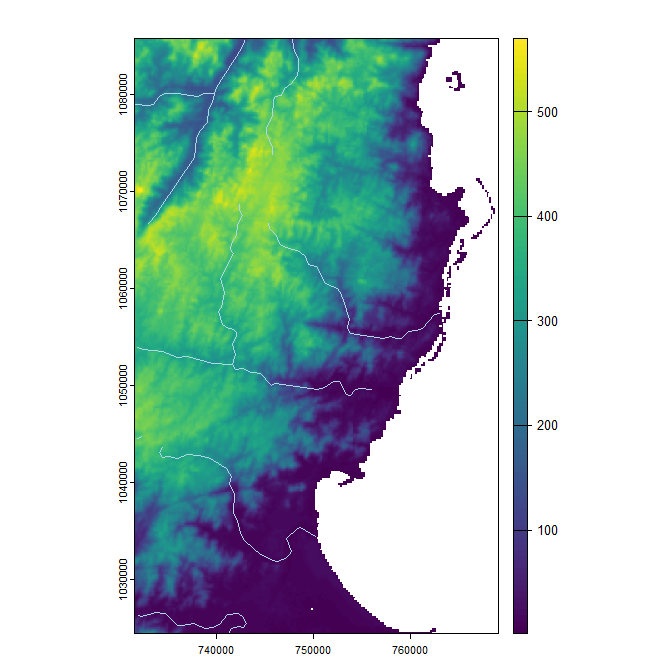
Before you use nearly any function in the package, you need to tell
fasterRaster where GRASS is installed on your
system. The installation folder will vary by operating system and maybe
GRASS version, but will look something like this:
# Choose the appropriate one, and modify as needed:
grassDir <- "C:/Program Files/GRASS GIS 8.4" # Windows
grassDir <- "/Applications/GRASS-8.4.app/Contents/Resources" # Mac OS
grassDir <- "/usr/local/grass" # LinuxNow, use the faster() function to tell
fasterRaster where GRASS is installed:
faster(grassDir = grassDir)The fast() function is the key function for loading a
raster or vector into fasterRaster format. Rasters in this
package are called GRasters and vectors
GVectors (the “G” stands for GRASS). We will
now convert the madElev raster, which is a
SpatRaster from the terra package, into a
GRaster.
elev <- fast(madElev)
elevYou will see the GRasters metadata:
class : GRaster
topology : 2D
dimensions : 1024, 626, NA, 1 (nrow, ncol, ndepth, nlyr)
resolution : 59.85157, 59.85157, NA (x, y, z)
extent : 731581.552, 769048.635, 1024437.272, 1085725.279 (xmin, xmax, ymin, ymax)
coord ref. : Tananarive (Paris) / Laborde Grid
name(s) : madElev
datatype : integer
min. value : 1
max. value : 570Next, we’ll do the same for the rivers vector. In this case, the
vector is an sf object from the sf package,
but we could also use a SpatVector from the
terra package.
rivers <- fast(madRivers)
riversclass : GVector
geometry : 2D lines
dimensions : 11, 11, 5 (geometries, sub-geometries, columns)
extent : 731627.1, 762990.132, 1024541.235, 1085580.454 (xmin, xmax, ymin, ymax)
coord ref. : Tananarive (Paris) / Laborde Grid
names : F_CODE_DES HYC_DESCRI NAM ISO NAME_0
type : <chr> <chr> <chr> <chr> <chr>
values : River/Stream Perennial/Permanent MANANARA MDG Madagascar
River/Stream Perennial/Permanent MANANARA MDG Madagascar
River/Stream Perennial/Permanent UNK MDG Madagascar
...and 8 more rows
Now, let’s add a 1000-m buffer to the rivers using
buffer(). As much as possible, fasterRaster
functions have the same names and same arguments as their counterparts
in the terra package to help users who are familiar with
that package.
Note, though, that the output from fasterRaster is not
necessarily guaranteed to be the same as output from the respective
functions terra. This is because there are different
methods to do the same thing, and the developers of GRASS
may have chosen different methods than the developers of other GIS
packages.
# width in meters because CRS is projected
river_buffers <- buffer(rivers, width = 1000)Now, let’s calculate the distances between the buffered areas and all
cells on the raster map using distance().
dist_to_rivers_meters <- distance(elev, river_buffers)Finally, let’s plot the output.
plot(dist_to_rivers_meters)
plot(river_buffers, border = 'white', add = TRUE)
plot(rivers, col = "lightblue", add = TRUE)
And that’s how it’s done! You can do almost anything in
fasterRaster you can do with terra. The
examples above do not show the advantage of fasterRaster
because the they do not use in large-in-memory/large-on-disk spatial
datasets. For very large datasets, fasterRaster can be much
faster! For example, for a large raster (many cells), the
distance() function in terra can take many
days to run and even crash R, whereas in
fasterRaster, it could take just a few minutes or
hours.
GRasters and GVectors from a
GRASS sessionYou can convert a GRaster to a SpatRaster
raster using rast():
terra_elev <- rast(elev)
To convert a GVector to the terra package’s
SpatVector, use vect():
terra_rivers <- vect(rivers)You can use writeRaster() and writeVector()
to save fasterRaster rasters and vectors directly to disk.
This will always be faster than using rast() or
vect() and then saving.
elev_temp_file <- tempfile(fileext = ".tif") # save as GeoTIFF
writeRaster(elev, elev_temp_file)
vect_temp_shp <- tempfile(fileext = ".shp") # save as shapefile
vect_temp_gpkg <- tempfile(fileext = ".gpkg") # save as GeoPackage
writeVector(rivers, vect_temp_shp)
writeVector(rivers, vect_temp_gpkg)fasterRaster versions will look something like
8.3.1.2, or more generally, M1.M2.S1.S2. Here,
M1.M2 will mirror the version of GRASS for
which fasterRaster was built and tested. For example,
fasterRaster version 8.4.x.x will work using
GRASS 8.4 (and version 8.3). The values in
S1.S2 refer to “major” and “minor” versions of
fasterRaster. That is, a change in the value of
S1 (e.g., from x.x.1.0 to
x.x.2.0) indicates changes that potentially break older
code developed with a prior version of fasterRaster. A
change in S2 refers to a bug fix, additional functionality
in an existing function, or the addition of an entirely new
function.
Note that the M1.M2 and S1.S2 increment
independently. For example, if the version changes from
8.3.1.5 to 8.4.1.5, then the new version has
been tested on GRASS 8.4, but code developed with version
8.3.1.x of fasterRaster should still work.
NOTE: While fasterRaster is still in
beta/alpha release, the version will look something like
8.3.0.7XXX, following Hadley Wickham’s guidelines for
versioning under development.
terra
package and Edzer Pebesma’s sf package
are good places to start if you are not familiar with doing GIS in
R.rgrass
package allows users to call any GRASS function with all of
its functionality, which in some cases is far beyond what is allowed by
fasterRaster.GRASS
functions used in this package and more.GRASS in R or R in
GRASS will help you to become a power-user of
GRASS in R.A publication is forthcoming! In the meantime, please see and cite:
fasterRaster: GIS in R
using GRASS for large vectors and rasters. EarthArXiv
preprint doi: <href =
“https://doi.org/10.31223/X52R0M”>10.31223/X52R0MAdam