The goal of dataspice is to make it easier for
researchers to create basic, lightweight, and concise metadata files for
their datasets by editing the kind of files they’re probably most
familiar with: CSVs. To spice up their data with a dash of metadata.
These metadata files can then be used to:
Metadata fields are based on Schema.org/Dataset and other metadata standards and represent a lowest common denominator which means converting between formats should be relatively straightforward.
An basic example repository for demonstrating what using
dataspice might look like can be found at https://github.com/amoeba/dataspice-example.
From there, you can also check out a preview of the HTML
dataspice generates at https://amoeba.github.io/dataspice-example
and how Google sees it at https://search.google.com/test/rich-results?url=https%3A%2F%2Famoeba.github.io%2Fdataspice-example%2F.
A much more detailed example has been created by Anna Krystalli at https://annakrystalli.me/dataspice-tutorial/ (GitHub repo).
You can install the latest version from CRAN:
install.packages("dataspice")create_spice()
# Then fill in template CSV files, more on this below
write_spice()
build_site() # Optional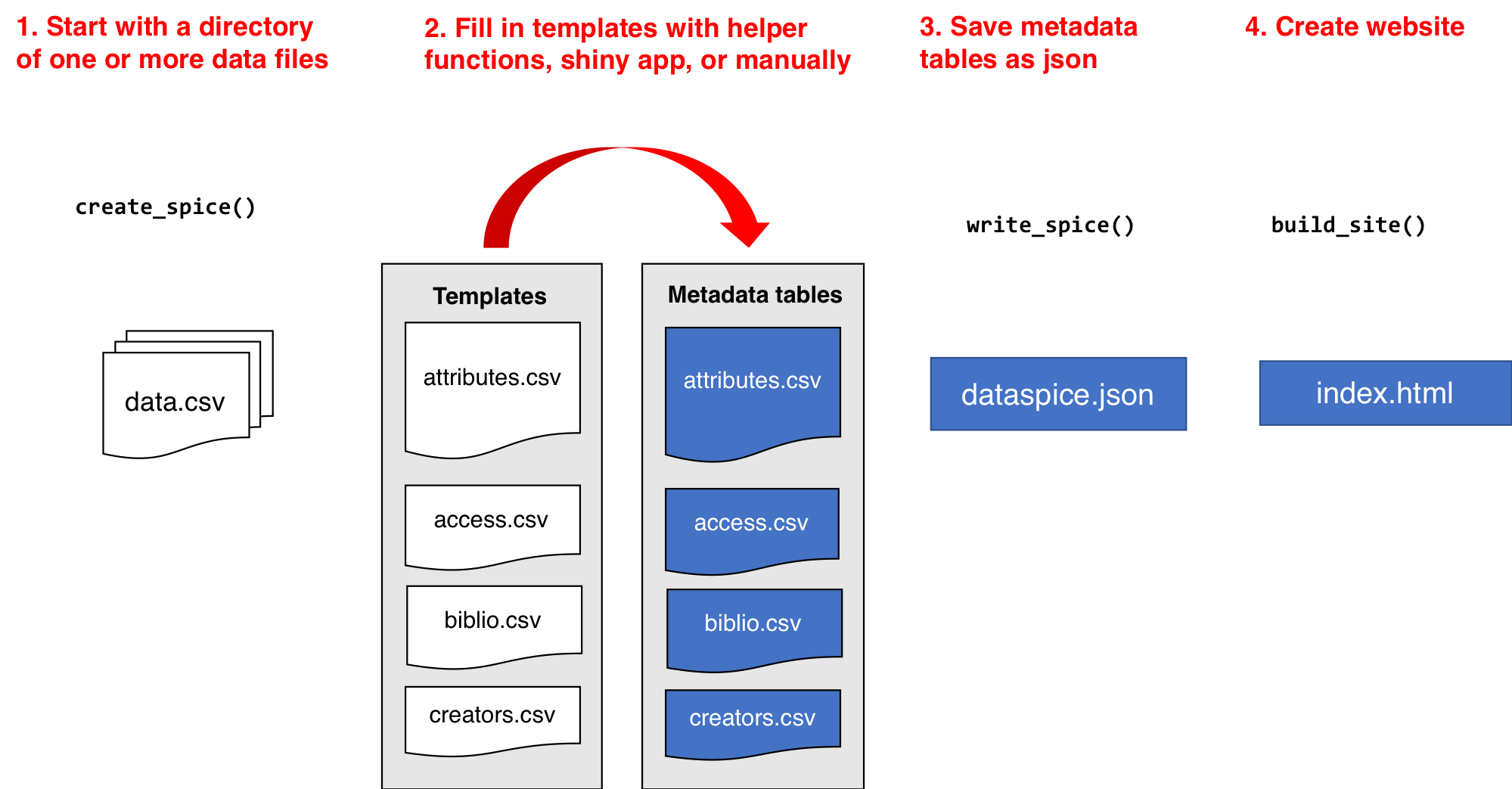
create_spice() creates template metadata spreadsheets in
a folder (by default created in the data folder in the
current working directory).
The template files are:
The user needs to fill in the details of the four template files. These csv files can be directly modified, or they can be edited using either the associated helper function and/or Shiny app.
prep_attributes() populates the
fileName and
variableName columns of the
attributes.csv file using the header row of the data
files.
prep_access() populates the
fileName,
name and
encodingFormat columns of the
access.csv file from the files in the folder containing the
data.
To see an example of how prep_attributes() works, load
the data files that ship with the package:
data_files <- list.files(system.file("example-dataset/", package = "dataspice"),
pattern = ".csv",
full.names = TRUE
)This function assumes that the metadata templates are in a folder
called metadata within a data folder.
attributes_path <- file.path("data", "metadata", "attributes.csv")Using purrr::map(), this function can be applied over
multiple files to populate the header names
data_files %>%
purrr::map(~ prep_attributes(.x, attributes_path),
attributes_path = attributes_path
)The output of prep_attributes() has the first two
columns filled out:
| fileName | variableName | description | unitText |
|---|---|---|---|
| BroodTables.csv | Stock.ID | NA | NA |
| BroodTables.csv | Species | NA | NA |
| BroodTables.csv | Stock | NA | NA |
| BroodTables.csv | Ocean.Region | NA | NA |
| BroodTables.csv | Region | NA | NA |
| BroodTables.csv | Sub.Region | NA | NA |
Each of the metadata templates can be edited interactively using a Shiny app:
edit_attributes() opens a Shiny app that can be used to
edit attributes.csv. The Shiny app displays the current
attributes table and lets the user fill in an informative
description and units (e.g. meters, hectares, etc.) for each
variable.edit_access() opens an editable version of
access.csvedit_creators() opens an editable version of
creators.csvedit_biblio() opens an editable version of
biblio.csv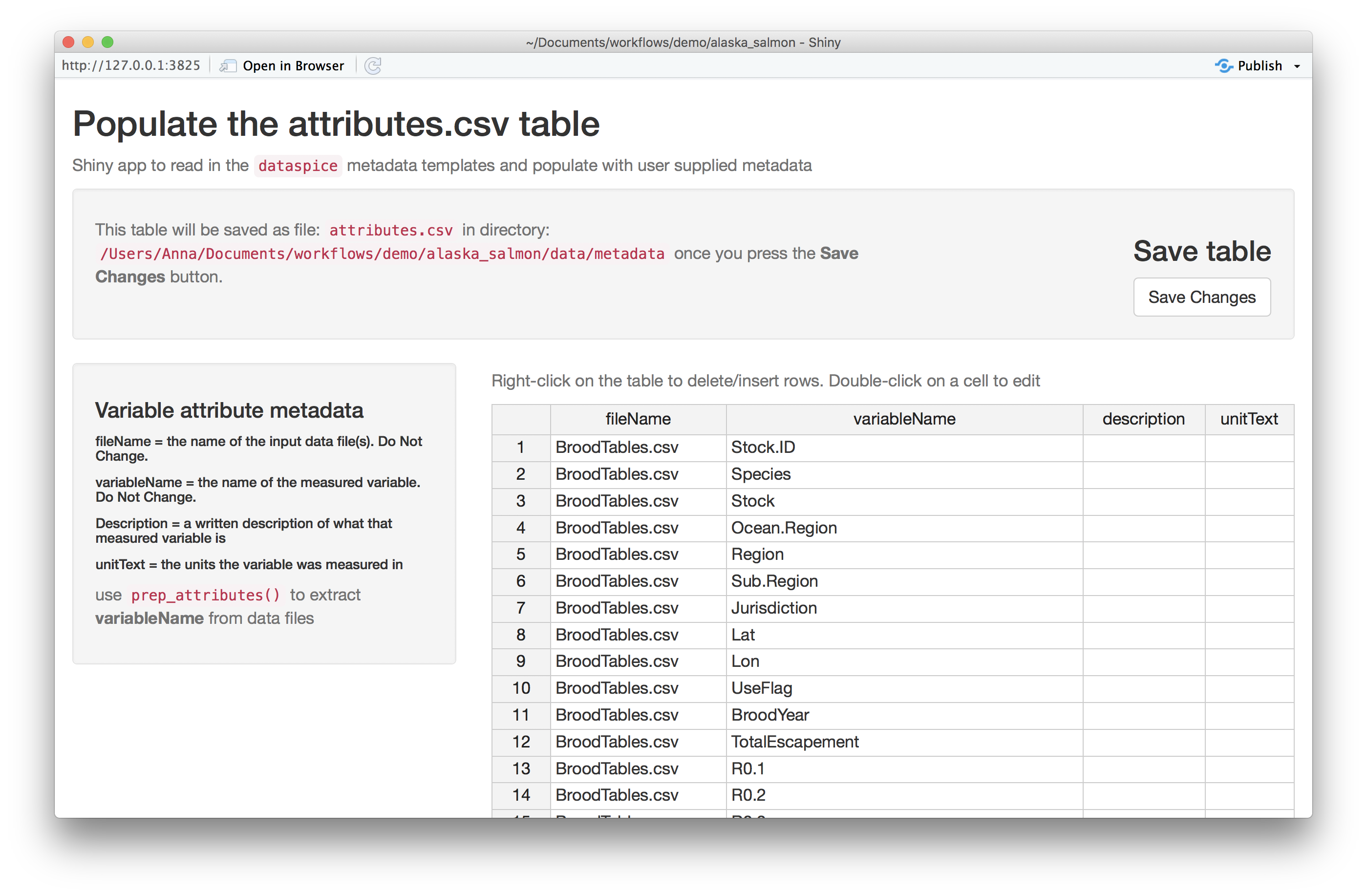
Remember to click on Save when finished editing.
The first few rows of the completed metadata tables in this example will look like this:
access.csv has one row for each file
| fileName | name | contentUrl | encodingFormat |
|---|---|---|---|
| StockInfo.csv | StockInfo.csv | NA | CSV |
| BroodTables.csv | BroodTables.csv | NA | CSV |
| SourceInfo.csv | SourceInfo.csv | NA | CSV |
attributes.csv has one row for each variable in each
file
| fileName | variableName | description | unitText |
|---|---|---|---|
| BroodTables.csv | Stock.ID | Unique stock identifier | NA |
| BroodTables.csv | Species | species of stock | NA |
| BroodTables.csv | Stock | Stock name, generally river where stock is found | NA |
| BroodTables.csv | Ocean.Region | Ocean region | NA |
| BroodTables.csv | Region | Region of stock | NA |
| BroodTables.csv | Sub.Region | Sub.Region of stock | NA |
biblio.csv is one row containing descriptors including
spatial and temporal coverage
| title | description | datePublished | citation | keywords | license | funder | geographicDescription | northBoundCoord | eastBoundCoord | southBoundCoord | westBoundCoord | wktString | startDate | endDate |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compiled annual statewide Alaskan salmon escapement counts, 1921-2017 | The number of mature salmon migrating from the marine environment to freshwater streams is defined as escapement. Escapement data are the enumeration of these migrating fish as they pass upstream, … | 2018-02-12 08:00:00 | NA | salmon, alaska, escapement | NA | NA | NA | 78 | -131 | 47 | -171 | NA | 1921-01-01 08:00:00 | 2017-01-01 08:00:00 |
creators.csv has one row for each of the dataset
authors
| id | name | affiliation | |
|---|---|---|---|
| NA | Jeanette Clark | National Center for Ecological Analysis and Synthesis | jclark@nceas.ucsb.edu |
| NA | Rich,Brenner | Alaska Department of Fish and Game | richard.brenner.alaska.gov |
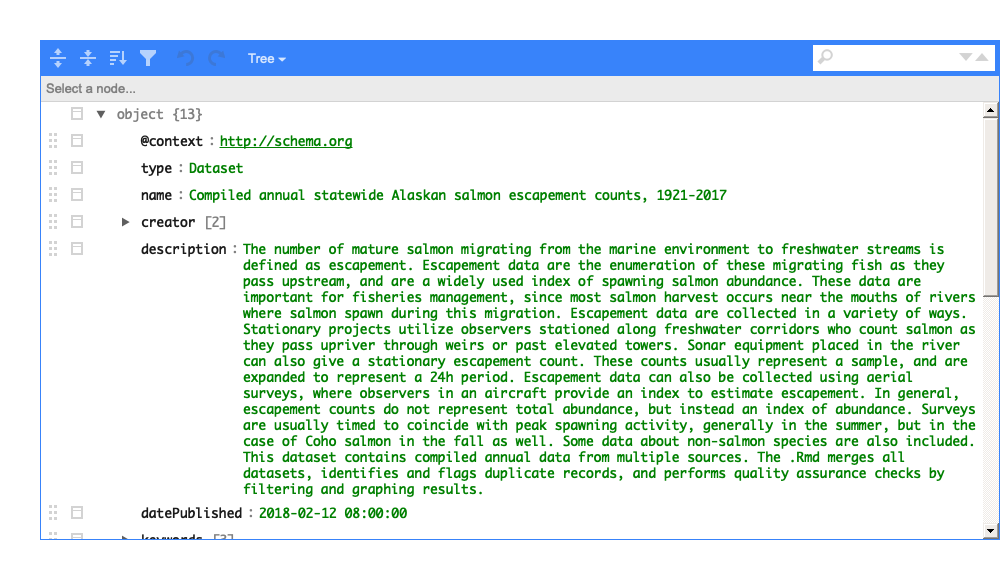
write_spice() generates a json-ld file (“linked data”)
to aid in dataset
discovery, creation of more extensive metadata (e.g. EML), and creating a
website.
Here’s a view of the dataspice.json file of the example
data:
build_site() creates a bare-bones
index.html file in the repository docs folder
with a simple view of the dataset with the metadata and an interactive
map. For example, this repository
results in this website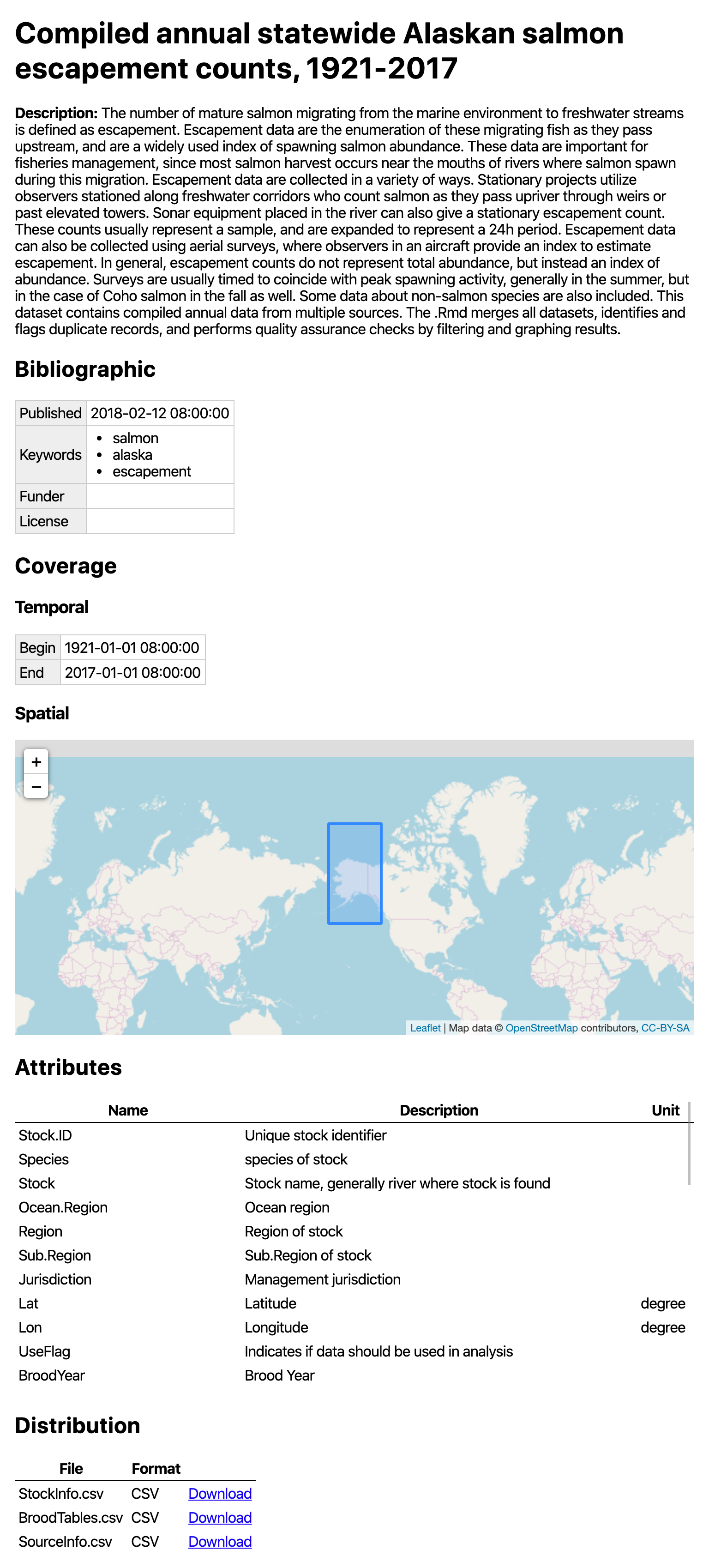
The metadata fields dataspice uses are based largely on
their compatibility with terms from Schema.org. However,
dataspice metadata can be converted to Ecological Metadata
Language (EML), a much richer schema. The conversion isn’t perfect but
dataspice will do its best to convert your
dataspice metadata to EML:
library(dataspice)
# Load an example dataspice JSON that comes installed with the package
spice <- system.file(
"examples", "annual-escapement.json",
package = "dataspice"
)
# Convert it to EML
eml_doc <- spice_to_eml(spice)
#> Warning: variableMeasured not crosswalked to EML because we don't have enough
#> information. Use `crosswalk_variables` to create the start of an EML attributes
#> table. See ?crosswalk_variables for help.
#> You might want to run EML::eml_validate on the result at this point and fix what validations errors are produced. You will commonly need to set `packageId`, `system`, and provide `attributeList` elements for each `dataTable`.You may receive warnings depending on which dataspice
fields you filled in and this process will very likely produce an
invalid EML record which is totally fine:
library(EML)
#>
#> Attaching package: 'EML'
#> The following object is masked from 'package:magrittr':
#>
#> set_attributes
eml_validate(eml_doc)
#> [1] FALSE
#> attr(,"errors")
#> [1] "Element '{https://eml.ecoinformatics.org/eml-2.2.0}eml': The attribute 'packageId' is required but missing."
#> [2] "Element '{https://eml.ecoinformatics.org/eml-2.2.0}eml': The attribute 'system' is required but missing."
#> [3] "Element 'dataTable': Missing child element(s). Expected is one of ( physical, coverage, methods, additionalInfo, annotation, attributeList )."
#> [4] "Element 'dataTable': Missing child element(s). Expected is one of ( physical, coverage, methods, additionalInfo, annotation, attributeList )."
#> [5] "Element 'dataTable': Missing child element(s). Expected is one of ( physical, coverage, methods, additionalInfo, annotation, attributeList )."This is because some fields in dataspice store
information in different structures and because EML requires many fields
that dataspice doesn’t have fields for. At this point, you
should look over the validation errors produced by
EML::eml_validate and fix those. Note that this will likely
require familiarity with the EML Schema and the EML package.
Once you’re done, you can write out an EML XML file:
out_path <- tempfile()
write_eml(eml_doc, out_path)
#> NULLLike converting dataspice to EML, we can convert an
existing EML record to a set of dataspice metadata tables
which we can then work from within dataspice:
library(EML)
eml_path <- system.file("example-dataset/broodTable_metadata.xml", package = "dataspice")
eml <- read_eml(eml_path)# Creates four CSVs files in the `data/metadata` directory
my_spice <- eml_to_spice(eml, "data/metadata")A few existing tools & data standards to help users in specific domains:
EML)…And others indexed in Fairsharing.org & the RDA metadata directory.
Please note that this package is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
This package was developed at rOpenSci’s 2018 unconf by (in alphabetical order):