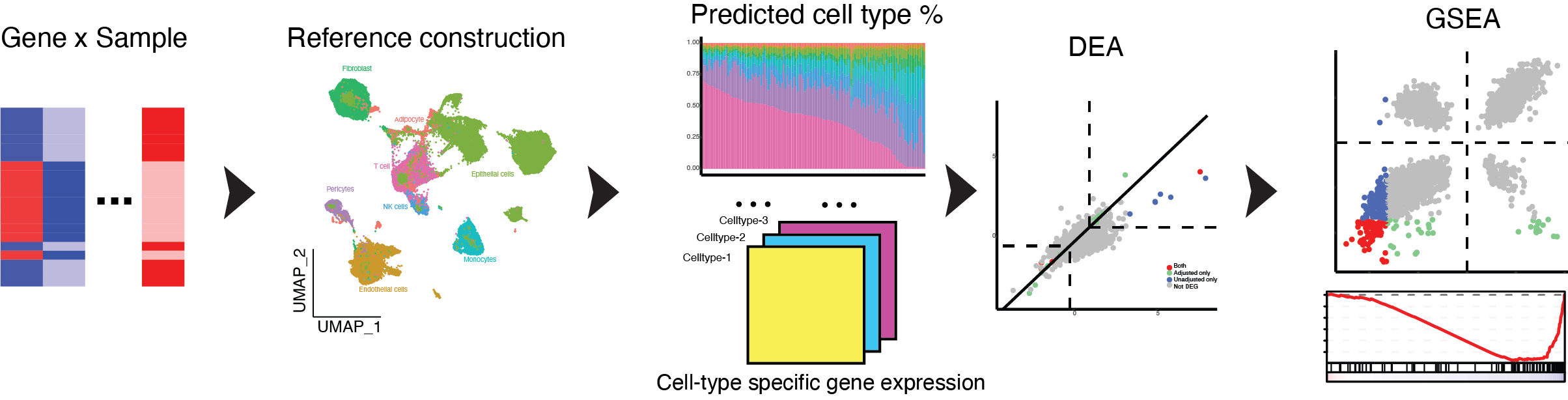

SCdeconR aims to provide a streamlined workflow from deconvolution of bulk RNA-seq data to downstream differential and gene-set enrichment analysis. SCdeconR provides a simulation framework to generate artificial bulk samples for benchmarking purposes. It also provides various visualization options to compare the influence of adjusting for cell-proportions differences on differential expression and pathway analyses.
# install devtools if it's not installed already
if (!require("devtools", quietly = TRUE)) install.packages("devtools")
devtools::install_github("liuy12/SCdeconR")To use scaden within SCdeconR, follow the below steps:
# install reticulate package first
install.packages("reticulate")Intall scaden python package:
Use pip:
pip install scaden
Or use Conda:
conda install scaden
Then provide your desired python path (that have scaden installed) to
option pythonpath for function scdecon. You
should be good to go.
The following packages are optional, and only needed for specific methods within SCdeconR.
# install BiocManager if it's not installed already
if (!require("BiocManager", quietly = TRUE)) install.packages("BiocManager")
# data normalization
## scater
BiocManager::install("scater")
## scran
BiocManager::install("scran")
## Linnorm
BiocManager::install("Linnorm")
## SingleCellExperiment
BiocManager::install("SingleCellExperiment")
# deconvolution methods
## FARDEEP
install.packages("FARDEEP")
## nnls
install.packages("nnls")
## MuSiC
devtools::install_github('xuranw/MuSiC')
## SCDC
devtools::install_github("meichendong/SCDC")
# differential expression
## DESeq2
BiocManager::install("DESeq2")
# cell-type specific gene expression
## spacexr
devtools::install_github("dmcable/spacexr", build_vignettes = FALSE)
# interactive plot
install.packages("plotly")library(SCdeconR)See here for detailed documentation and tutorials.
See here for a document to reproduce the results from the study.