
The goal of SANple is to estimate Bayesian nested mixture models via MCMC methods. Specifically, the package implements the common atoms model (Denti et al., 2023), hybrid finite-infinite models (D’Angelo and Denti, 2024). All models use Gaussian mixtures with a normal-inverse-gamma prior distribution on the parameters. Additional functions are provided to help analyzing the results of the fitting procedure.
You can install the development version of SANple from GitHub with:
# install.packages("devtools")
devtools::install_github("laura-dangelo/SANple")This is a basic example which shows you how to solve a common problem:
library(SANple)
#> Loading required package: scales
#> Loading required package: RColorBrewer
## basic example code
set.seed(123)
y <- c(rnorm(50,-5,1), rnorm(170,0,1),rnorm(70,5,1))
g <- c(rep(1,150), rep(2, 140))
plot(density(y[g==1]), xlim = c(-10,10), main = "", xlab = "")
lines(density(y[g==2]), col = "cyan4")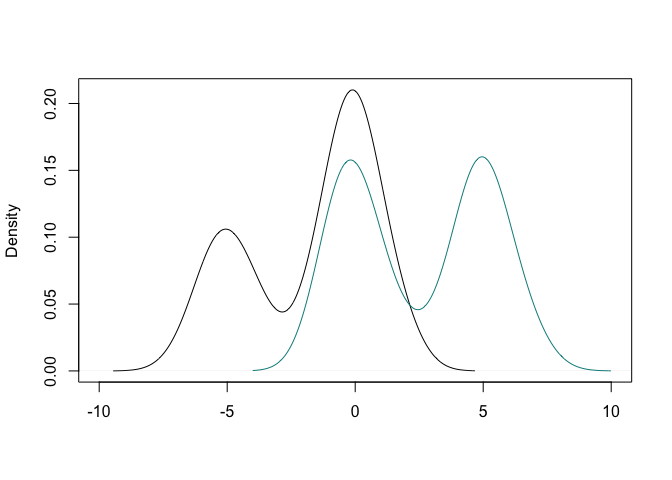
out <- sample_fiSAN(nrep = 3000, burn = 1000, y = y, group = g, beta = 0.01)
out
#>
#> MCMC result of fiSAN model
#> -----------------------------------------------
#> Model estimated on 290 total observations and 2 groups
#> Total MCMC iterations: 3000
#> maxL: 50 - maxK: 50
#> Elapsed time: 3.374 secs
#>
clusters <- estimate_clusters(out)
clusters
#> Summary of the estimated observational and distributional clusters
#>
#> ----------------------------------
#> Estimated number of observational clusters: 4
#> Estimated number of distributional clusters: 2
#> ----------------------------------
#>
#> Distributional cluster 1
#> post_mean post_var
#> 1 -4.96559645 0.8572352
#> 2 -0.05374608 0.9306906
#>
#> Distributional cluster 2
#> post_mean post_var
#> 3 -0.00160923 0.7737203
#> 4 5.03452815 0.8314760
#>
plot(out, estimated_clusters = clusters)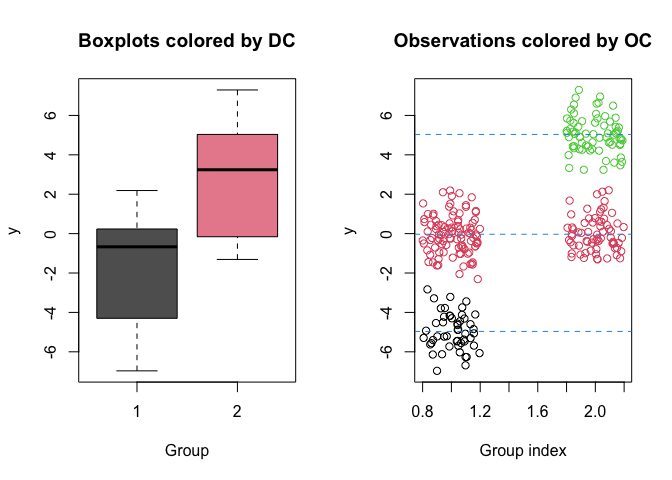
D’Angelo, L., and Denti, F. (2024). A Finite-Infinite Shared Atoms Nested Model for the Bayesian Analysis of Large Grouped Data Sets. Bayesian Analysis
Denti, F., Camerlenghi, F., Guindani, M., Mira, A., 2023. A Common Atoms Model for the Bayesian Nonparametric Analysis of Nested Data. Journal of the American Statistical Association. 118(541), 405–416.