You can install CodelistGenerator from CRAN
install.packages("CodelistGenerator")Or you can also install the development version of CodelistGenerator
install.packages("remotes")
remotes::install_github("darwin-eu/CodelistGenerator")library(dplyr)
library(CDMConnector)
library(CodelistGenerator)For this example we’ll use the Eunomia dataset (which only contains a subset of the OMOP CDM vocabularies)
requireEunomia()
db <- DBI::dbConnect(duckdb::duckdb(), dbdir = eunomiaDir())
cdm <- cdmFromCon(db,
cdmSchema = "main",
writeSchema = "main",
writePrefix = "cg_")OMOP CDM vocabularies are frequently updated, and we can identify the version of the vocabulary of our Eunomia data
vocabularyVersion(cdm = cdm)
#> [1] "v5.0 18-JAN-19"CodelistGenerator provides functions to extract code lists based on vocabulary hierarchies. One example is `getDrugIngredientCodes, which we can use, for example, to get the concept IDs used to represent aspirin and diclofenac.
ing <- getDrugIngredientCodes(cdm = cdm,
name = c("aspirin", "diclofenac"),
nameStyle = "{concept_name}")
ing
#>
#> - aspirin (2 codes)
#> - diclofenac (1 codes)
ing$aspirin
#> [1] 1112807 19059056
ing$diclofenac
#> [1] 1124300CodelistGenerator can also support systematic searches of the vocabulary tables to support codelist development. A little like the process for a systematic review, the idea is that for a specified search strategy, CodelistGenerator will identify a set of concepts that may be relevant, with these then being screened to remove any irrelevant codes by clinical experts.
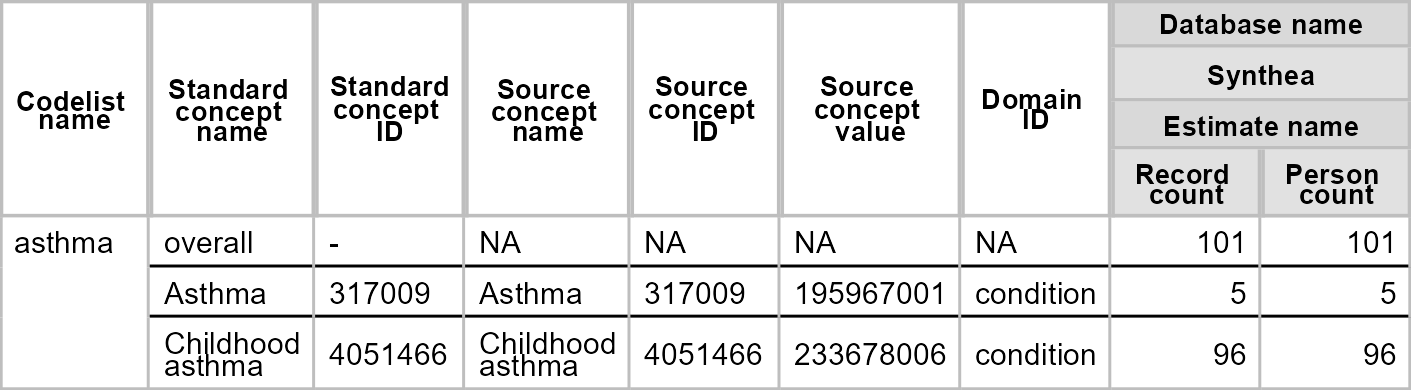
We can do a simple search for asthma
asthma_codes1 <- getCandidateCodes(
cdm = cdm,
keywords = "asthma",
domains = "Condition"
)
asthma_codes1 |>
glimpse()
#> Rows: 2
#> Columns: 6
#> $ concept_id <int> 317009, 4051466
#> $ found_from <chr> "From initial search", "From initial search"
#> $ concept_name <chr> "Asthma", "Childhood asthma"
#> $ domain_id <chr> "Condition", "Condition"
#> $ vocabulary_id <chr> "SNOMED", "SNOMED"
#> $ standard_concept <chr> "S", "S"But perhaps we want to exclude certain concepts as part of the search strategy, in this case we can add these like so
asthma_codes2 <- getCandidateCodes(
cdm = cdm,
keywords = "asthma",
exclude = "childhood",
domains = "Condition"
)
asthma_codes2 |>
glimpse()
#> Rows: 1
#> Columns: 6
#> $ concept_id <int> 317009
#> $ found_from <chr> "From initial search"
#> $ concept_name <chr> "Asthma"
#> $ domain_id <chr> "Condition"
#> $ vocabulary_id <chr> "SNOMED"
#> $ standard_concept <chr> "S"As well as functions for finding codes, we also have functions to summarise their use. Here for
library(flextable)
asthma_code_use <- summariseCodeUse(list("asthma" = asthma_codes1$concept_id) |>
newCodelist(),
cdm = cdm
)
tableCodeUse(asthma_code_use, type = "flextable", style = "darwin")